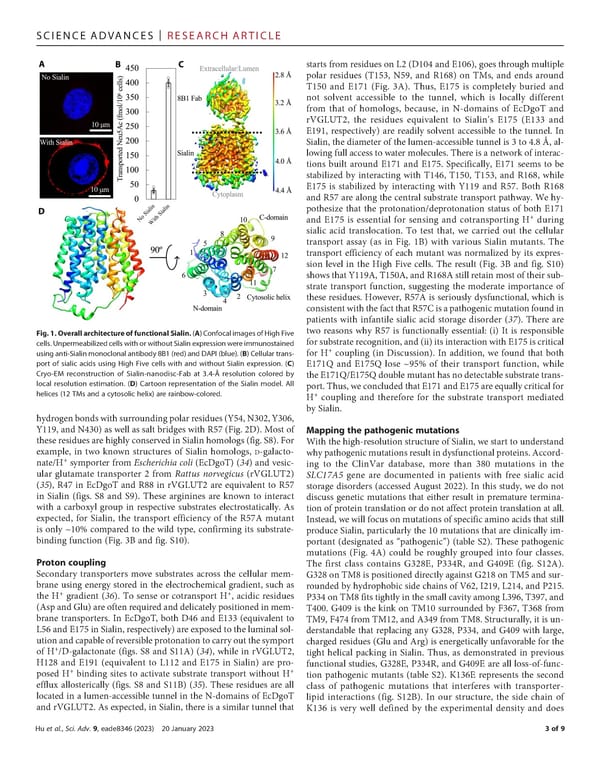
SCIENCEADVANCES | RESEARCHARTICLE starts from residues on L2 (D104 and E106), goes through multiple polar residues (T153, N59, and R168) on TMs, and ends around T150 and E171 (Fig. 3A). Thus, E175 is completely buried and not solvent accessible to the tunnel, which is locally different from that of homologs, because, in N-domains of EcDgoT and rVGLUT2, the residues equivalent to Sialin’s E175 (E133 and E191, respectively) are readily solvent accessible to the tunnel. In Sialin, the diameter of the lumen-accessible tunnel is 3 to 4.8 Å, al- lowing full access to water molecules. There is a network of interac- tions built around E171 and E175. Specifically, E171 seems to be stabilized by interacting with T146, T150, T153, and R168, while E175 is stabilized by interacting with Y119 and R57. Both R168 and R57 are along the central substrate transport pathway. We hy- pothesize that the protonation/deprotonation status of both E171 and E175 is essential for sensing and cotransporting H+ during sialic acid translocation. To test that, we carried out the cellular transport assay (as in Fig. 1B) with various Sialin mutants. The transport efficiency of each mutant was normalized by its expres- sion level in the High Five cells. The result (Fig. 3B and fig. S10) showsthatY119A,T150A,andR168Astillretainmostoftheirsub- strate transport function, suggesting the moderate importance of these residues. However, R57A is seriously dysfunctional, which is consistentwiththefactthatR57Cisapathogenicmutationfoundin patients with infantile sialic acid storage disorder (37). There are Fig.1.OverallarchitectureoffunctionalSialin.(A)ConfocalimagesofHighFive two reasons why R57 is functionally essential: (i) It is responsible cells. UnpermeabilizedcellswithorwithoutSialinexpressionwereimmunostained for substrate recognition, and (ii) its interaction with E175 is critical + usinganti-Sialin monoclonalantibody8B1(red)andDAPI(blue).(B)Cellulartrans- for H coupling (in Discussion). In addition, we found that both port of sialic acids using High Five cells with and without Sialin expression. (C) E171Q and E175Q lose ~95% of their transport function, while Cryo-EM reconstruction of Sialin-nanodisc-Fab at 3.4-Å resolution colored by theE171Q/E175Qdoublemutanthasnodetectablesubstratetrans- local resolution estimation. (D) Cartoon representation of the Sialin model. All port. Thus, we concludedthatE171andE175areequallycriticalfor helices (12 TMs and a cytosolic helix) are rainbow-colored. + H coupling and therefore for the substrate transport mediated by Sialin. hydrogenbondswithsurroundingpolarresidues(Y54,N302,Y306, Y119, and N430) as well as salt bridges with R57 (Fig. 2D). Most of Mappingthepathogenicmutations these residues are highly conserved in Sialin homologs (fig. S8). For Withthehigh-resolution structure of Sialin, we start to understand example, in two known structures of Sialin homologs, D-galacto- whypathogenicmutationsresultindysfunctionalproteins.Accord- + nate/H symporter from Escherichia coli (EcDgoT) (34) and vesic- ing to the ClinVar database, more than 380 mutations in the ular glutamate transporter 2 from Rattus norvegicus (rVGLUT2) SLC17A5 gene are documented in patients with free sialic acid (35), R47 in EcDgoT and R88 in rVGLUT2 are equivalent to R57 storage disorders (accessed August 2022). In this study, we do not in Sialin (figs. S8 and S9). These arginines are known to interact discuss genetic mutations that either result in premature termina- with a carboxyl group in respective substrates electrostatically. As tion of protein translation or do not affect protein translation at all. expected, for Sialin, the transport efficiency of the R57A mutant Instead, we will focus on mutations of specific amino acids that still is only ~10% compared to the wild type, confirming its substrate- produce Sialin, particularly the 10 mutations that are clinically im- binding function (Fig. 3B and fig. S10). portant (designated as “pathogenic”) (table S2). These pathogenic mutations (Fig. 4A) could be roughly grouped into four classes. Proton coupling The first class contains G328E, P334R, and G409E (fig. S12A). Secondary transporters move substrates across the cellular mem- G328onTM8ispositioneddirectlyagainstG218onTM5andsur- brane using energy stored in the electrochemical gradient, such as rounded by hydrophobic side chains of V62, I219, L214, and P215. + + the H gradient (36). To sense or cotransport H , acidic residues P334onTM8fitstightlyinthesmallcavityamongL396,T397,and (AspandGlu)areoftenrequiredanddelicatelypositionedinmem- T400. G409 is the kink on TM10 surrounded by F367, T368 from brane transporters. In EcDgoT, both D46 and E133 (equivalent to TM9,F474fromTM12,andA349fromTM8.Structurally,itisun- L56andE175inSialin,respectively)areexposedtotheluminalsol- derstandable that replacing any G328, P334, and G409 with large, utionandcapableofreversibleprotonationtocarryoutthesymport charged residues (Glu and Arg) is energetically unfavorable for the + of H /D-galactonate (figs. S8 and S11A) (34), while in rVGLUT2, tight helical packing in Sialin. Thus, as demonstrated in previous H128 and E191 (equivalent to L112 and E175 in Sialin) are pro- functional studies, G328E, P334R, and G409E are all loss-of-func- + + posed H binding sites to activate substrate transport without H tion pathogenic mutants (table S2). K136E represents the second efflux allosterically (figs. S8 and S11B) (35). These residues are all class of pathogenic mutations that interferes with transporter- located in a lumen-accessible tunnel in the N-domains of EcDgoT lipid interactions (fig. S12B). In our structure, the side chain of and rVGLUT2.Asexpected, in Sialin, there is a similar tunnel that K136 is very well defined by the experimental density and does Huetal., Sci. Adv. 9, eade8346 (2023) 20 January 2023 3of9
 The molecular mechanism of sialic acid transport mediated by Sialin Page 2 Page 4
The molecular mechanism of sialic acid transport mediated by Sialin Page 2 Page 4